SickKids scientists discover novel genetic contributors to autism
Summary:
Breakthrough SickKids study of thousands of repetitive DNA “wrinkles” identifies previously hidden genetic alterations for the first time.
TORONTO - New research from The Hospital for Sick Children (SickKids) significantly advances our understanding of the human genome and promises to have immediate impact for individuals and their families with autism spectrum disorder (ASD).
ASD refers to a group of neurodevelopmental conditions resulting in challenges related to communication, social understanding and behaviour. Current research estimates that genetic factors should be found in anywhere from 50 to 90 per cent of individuals with ASD. Yet, scientists are only able to identify the genetic factor underlying autism for less than 20 per cent of people.
Now, a research team led by Dr. Ryan Yuen, Scientist in the Genetics & Genome Biology program at SickKids, has moved the needle, uncovering new genetic contributors for autism. Their findings were published online on July 27, 2020, in Nature.
DNA tandem repeat expansions: Why more isn’t always better
The research team wanted to look at the implications of tandem repeats in the DNA of people with ASD. A tandem repeat occurs when nucleotides, known as the building blocks of DNA, are repeated adjacently two or more times. These repeats are like DNA “wrinkles” and the larger they become, the greater the likelihood that errors in gene function will result.
Similar to how photocopying a piece of wrinkled paper will sometimes result in more wrinkles, when a strand of DNA with a tandem repeat replicates, the repeat often grows longer. These are called tandem repeat expansions and depending on their length and location in the genome, they can result in gene dysregulation, contributing to conditions such as Fragile X syndrome and Huntington’s Disease.
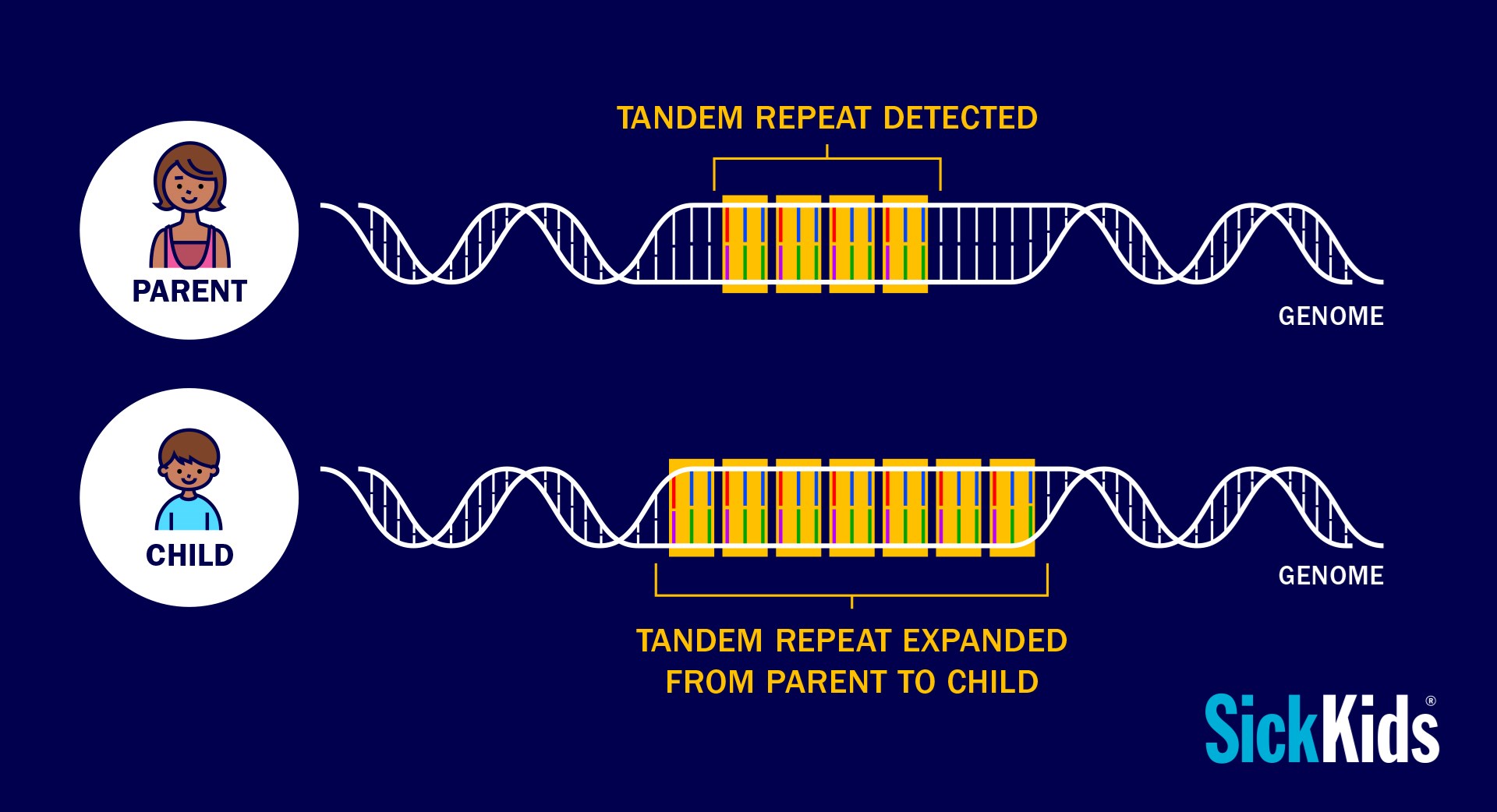
Previously, scientists could only search for tandem repeats in one gene at a time and were faced with challenges in determining whether a tandem repeat was long enough to impact gene function. With close to a million tandem repeats in the genome, looking for tandem repeats that contribute meaningfully to ASD would be like looking for a needle in a haystack.
Yuen’s team developed an analytic approach that could search and find significant tandem repeat expansions across the entire genome. “We’re effectively searching millions of repetitive regions of DNA simultaneously and decoding them in a way no one has ever been able to do,” says Yuen, Principal Investigator of the study and an Assistant Professor in the Department of Molecular Genetics at the University of Toronto.
“Our computational approach can determine the outliers – the tandem repeats that have an exceptionally high number of repetitions and could change how our genes function.”
A leap forward for ASD research
The researchers applied their method to the whole genome sequencing data of over 5,000 families with ASD and more than 4,000 individuals from the general population. They found 2,588 different places in the genome where tandem repeat expansions associated with particular genes were much more prevalent among individuals with ASD compared to those without ASD. This discovery shows tandem repeat expansions are as great a contributor to ASD as the previous major contributing genetic factor (de novo genetic variants).
The researchers found some of the tandem repeat expansions occurred in genes previously known to be implicated in ASD. However, about 50 per cent occurred in entirely new genes that were never thought to be involved in ASD, including ones that are involved in some neuromuscular disorders. Furthermore, the location of a tandem repeat expansion was associated with certain characteristics and behaviours such as IQ and life skills.
“In order to conduct a robust analysis, we drew upon Autism Speaks’ MSSNG, a growing whole genome sequence database of families affected by ASD who are keen to better understand the many questions we all have about the condition,” says Dr. Stephen Scherer, co-author of the study, Director of The Centre for Applied Genomics (TCAG) at SickKids, and Director of the McLaughlin Centre at the University of Toronto. “This discovery would not have been possible without the families who are as committed to scientific exploration as the researchers themselves.”
Novel approach could uncover new therapeutic targets for wide range of neurodevelopmental disorders
The research team stresses that while they investigated the feasibility of their approach by detecting tandem repeat expansions for individuals with ASD, it can likely be applied to many different neurodevelopmental disorders. Their approach opens new opportunities for genome characterization by providing all researchers with a tool that can be applied to almost any genetic condition.
“By applying our method to a condition as complex as ASD, which can have a wide variety of outcomes from one person to another with different comorbidities, we’ve shown our approach can effectively detect tandem repeat expansions, no matter the clinical presentation. It is crucial to examine the entire genome to fully understand how genetics contribute to these complex conditions,” says Yuen.
“Discovering the impact of tandem repeat expansions and where they are located has major implications for the future of precision medicine in many neurodevelopmental conditions.”

In addition to conducting further investigations into the involvement of tandem repeat expansions in ASD, the research team is currently working on improving autism diagnoses by applying their approach to genomes of others with different clinical presentations. Yuen and his collaborators believe their novel approach could revolutionize the future of precision diagnostics.
This research was supported by the SickKids Catalyst Scholar program in Genetics, a Dataset Analysis Grant from Autism Speaks, an Accelerator Grant from the University of Toronto McLaughlin Centre, the NARSAD Young Investigator award, the Nancy E.T. Fahrner Award and SickKids Foundation. Other infrastructure, funding, and support for the generation and analysis of genomic data came from Autism Speaks (MSSNG Project), KRG Children's Charitable Foundation, Verily, Illumina, Mayo Clinic, the Canadian Institutes of Health Research (CIHR), the National Institutes of Health, the Ontario Brain Institute, Brain Canada, Kids Brain Health Network, Genome Canada/Ontario Genomics Institute, the Government of Ontario, the Canadian Institute for Advanced Research (CIFAR), and the Canada Foundation for Innovation (CFI).

